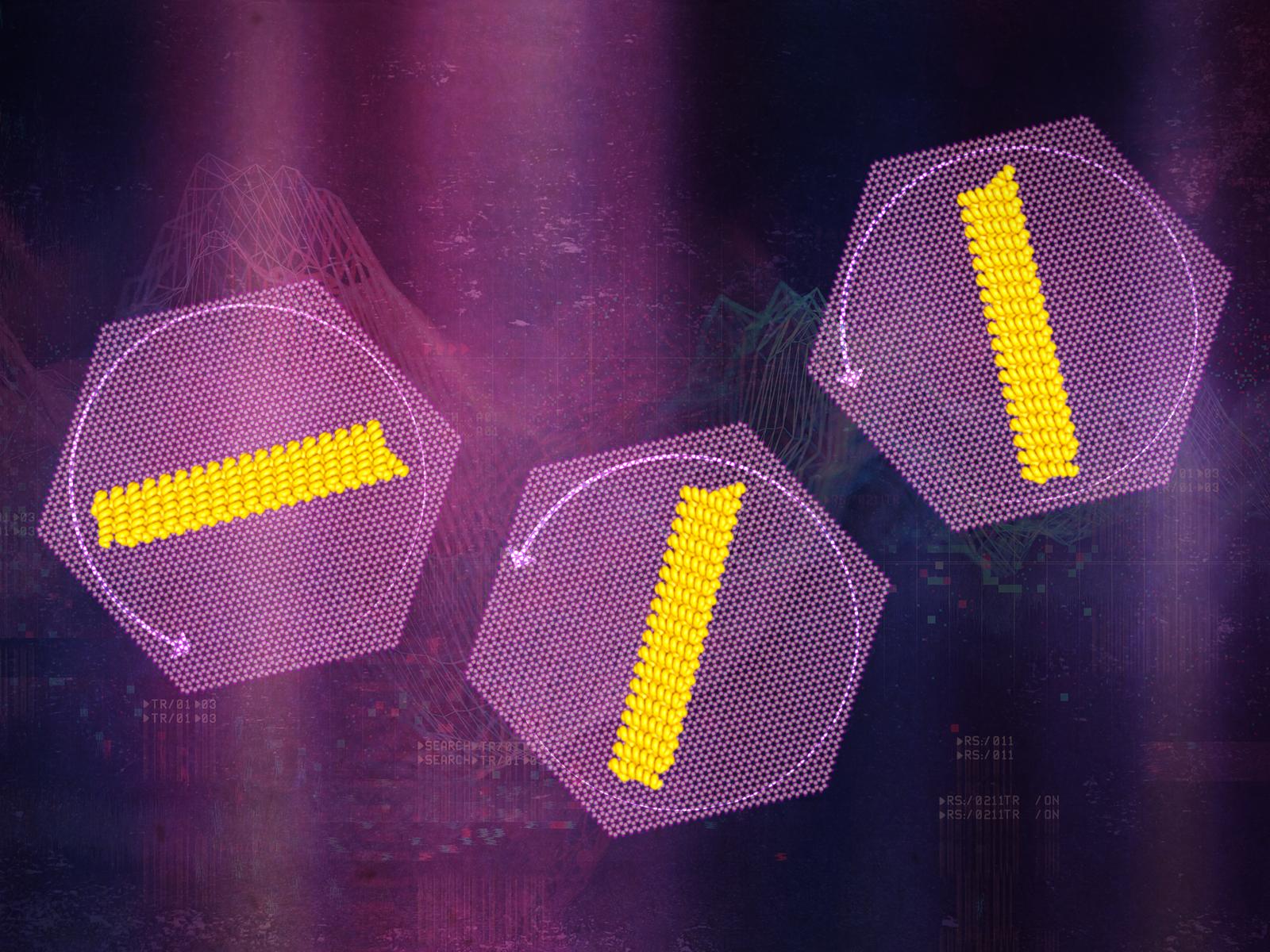
New analysis adopted proteins as they rotated on a mineral floor, figuring out surprising movement. Credit score: Illustration by Stephanie King | Pacific Northwest Nationwide Laboratory
Higher understanding of protein motion can facilitate superior supplies design
Organic supplies corresponding to bones, tooth, and seashells are extraordinarily sturdy. Their power stems from their composition, which consists of a mixture of exhausting rock-like minerals and resilient carbon-based compounds corresponding to proteins. Supplies scientists are drawing inspiration from organic supplies to develop a brand new technology of superior supplies comprised of proteins and minerals. Nevertheless, scientists should first know how proteins connect and assemble on mineral surfaces.
Proteins are a key kind of enormous, biologically related natural molecule, important to life on Earth. Along with naturally occurring proteins, researchers can customized create proteins with particular traits, buildings, and properties. This contains designing proteins that may connect to totally different surfaces, together with minerals like mica. Controlling and understanding protein attachment is central to assembling superior bio-inspired supplies.
A staff of researchers from Pacific Northwest Nationwide Laboratory (PNNL), the College of Washington (UW), and Lawrence Berkeley Nationwide Laboratory (Berkeley Lab) labored collectively to trace how specifically designed protein nanorods moved on a mica floor. Their findings had been lately printed within the Proceedings of the Nationwide Academy of Science. The staff created a sequence of different-sized protein nanorods particularly designed to bind to mica in partnership with the College of Washington’s Institute for Protein Design. The researchers then employed high-speed microscopy to see particular person nanorods rotate in real-time.
“We had been capable of observe the protein nanorods at unprecedented ranges of decision,” mentioned Shuai Zhang, a Analysis Assistant Professor within the Division of Supplies Science & Engineering at UW who has a joint appointment with PNNL. “The atomic pressure microscope we used is extremely highly effective, permitting us to see particular person molecule actions in real-time.”
To precisely observe protein rotation, the researchers needed to examine the protein-mica system in water. This surroundings mimics the situations of protein meeting on actual mineral surfaces.
Understanding totally different movement
Observing the system below a microscope produced huge portions of information. The sheer quantity of information made it difficult to investigate. The staff at Berkeley Lab solved that downside by creating a brand new machine-learning algorithm that dramatically decreased the time wanted to course of the pictures. From there, the researchers had been ready to take a look at how briskly the proteins moved and the way far they had been rotating per particular person transfer.
Their observations confirmed that the proteins principally behaved as anticipated, i.e., moved by making small jumps, following a mannequin of movement traceable again to Einstein. Nevertheless, the proteins sometimes made massive, fast jumps that the mannequin couldn’t clarify.
To unravel these several types of movement, the staff carried out simulations based mostly on the microscopy information. They discovered that the power of the protein-surface bond managed how the proteins may rotate. More often than not the proteins stay strongly certain to the mica floor, solely capable of make small motions. Often they seem to briefly detach from the mica. Throughout these brief intervals of time, the proteins can transfer rapidly in massive jumps.
“Evaluating our observational information and simulations allowed us to establish each forms of protein movement,” mentioned Ben Legg, a chemist at PNNL. “We predict that the big jumps have necessary penalties for assembling protein-mineral buildings.”
Understanding how particular person organic molecules transfer will help researchers develop higher strategies to assemble massive numbers of proteins on surfaces.
This work was funded by the Division of Vitality via the Heart for the Science of Synthesis Throughout Scales and the Scientific Discovery via Superior Computing (SciDAC) program. The PNNL analysis staff additionally included James De Yoreo. The UW staff included Harley Pyles and David Baker. The Berkeley Lab staff consisted of Robbie Sadre, Talita Perciano, E. Wes Bethe, and Oliver Rübel.
Reference: “Rotational dynamics and transition mechanisms of surface-adsorbed proteins” by Shuai Zhang, Robbie Sadre, Benjamin A. Legg, Harley Pyles, Talita Perciano, E. Wes Bethel, David Baker, Oliver Rübel and James J. De Yoreo, 11 April 2022, Proceedings of the Nationwide Academy of Science.
DOI: 10.1073/pnas.2020242119
Post a Comment