
Ocean Water Samples Yield Treasure Trove of RNA Virus Knowledge
Ocean water samples collected world wide have yielded a treasure trove of latest knowledge about RNA viruses, increasing ecological analysis potentialities and reshaping our understanding of how these small however important submicroscopic particles advanced.
Combining machine-learning analyses with conventional evolutionary timber, a world group of researchers has recognized 5,500 new RNA virus species that signify all 5 identified RNA virus phyla and counsel there are no less than 5 new RNA virus phyla wanted to seize them.
Probably the most considerable assortment of newly recognized species belong to a proposed phylum researchers named Taraviricota, a nod to the supply of the 35,000 water samples that enabled the evaluation: the Tara Oceans Consortium, an ongoing international research onboard the schooner Tara of the affect of local weather change on the world’s oceans.
“There’s a lot new variety right here – and a complete phylum, the Taraviricota,have been discovered everywhere in the oceans, which suggests they’re ecologically necessary,” mentioned lead writer Matthew Sullivan, professor of microbiology at The Ohio State College.
“RNA viruses are clearly necessary in our world, however we often solely research a tiny slice of them – the few hundred that hurt people, crops, and animals. We needed to systematically research them on a really large scale and discover an surroundings nobody had checked out deeply, and we acquired fortunate as a result of nearly each species was new, and plenty of have been actually new.”
The research was revealed on April 7, 2022, within the journal Science.
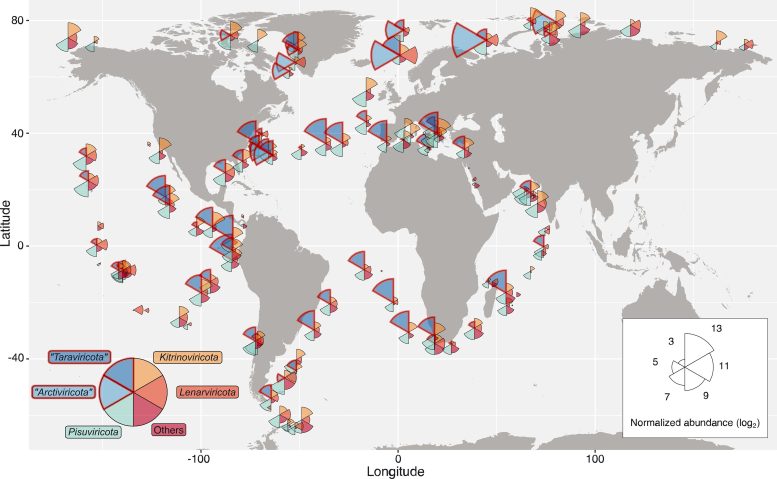
This map reveals the distribution of RNA viruses throughout the ocean. Wedge dimension is proportional to the common abundance of viruses current in that space, and wedge colour signifies virus phyla. Credit score: Reprinted with permission from Zayed et al., Science Quantity 376:156(2022)
Whereas microbes are important contributors to all life on the planet, viruses that infect or work together with them have a wide range of influences on microbial capabilities. Most of these viruses are believed to have three principal capabilities: killing cells, altering how contaminated cells handle vitality, and transferring genes from one host to a different.
Realizing extra about virus variety and abundance on the earth’s oceans will assist clarify marine microbes’ function in ocean adaptation to local weather change, the researchers say. Oceans take up half of the human-generated carbon dioxide from the ambiance, and earlier analysis by this group has recommended that marine viruses are the “knob” on a organic pump affecting how carbon within the ocean is saved.
By taking over the problem of classifying RNA viruses, the group entered waters nonetheless rippling from earlier taxonomy categorization efforts that targeted totally on RNA viral pathogens. Throughout the organic kingdom Orthornavirae, 5 phyla have been not too long ago acknowledged by the Worldwide Committee on Taxonomy of Viruses (ICTV).
“RdRp is meant to be one of the crucial historical genes – it existed earlier than there was a necessity for DNA. So we’re not simply tracing the origins of viruses, but in addition tracing the origins of life.” — Ahmed Zayed
Although the analysis group recognized a whole lot of latest RNA virus species that match into these present divisions, their evaluation recognized hundreds extra species that they clustered into 5 new proposed phyla: Taraviricota, Pomiviricota, Paraxenoviricota, Wamoviricota and Arctiviricota,which, like Taraviricota, options extremely considerable species – no less than in climate-critical Arctic Ocean waters, the world of the world the place warming circumstances wreak essentially the most havoc.
Sullivan’s group has lengthy cataloged DNA virus species within the oceans, rising the numbers from just a few thousand in 2015 and 2016 to 200,000 in 2019. For these research, scientists had entry to viral particles to finish the evaluation.
In these present efforts to detect RNA viruses, there have been no viral particles to check. As a substitute, researchers extracted sequences from genes expressed in organisms floating within the sea, and narrowed the evaluation to RNA sequences that contained a signature gene, known as RdRp, which has advanced for billions of years in RNA viruses, and is absent from different viruses or cells.
As a result of RdRp’s existence dates to when life was first detected on Earth, its sequence place has diverged many instances, that means conventional phylogenetic tree relationships have been unattainable to explain with sequences alone. As a substitute, the group used machine studying to arrange 44,000 new sequences in a method that would deal with these billions of years of sequence divergence, and validated the tactic by exhibiting the method might precisely classify sequences of RNA viruses already recognized.
“We needed to benchmark the identified to check the unknown,” mentioned Sullivan, additionally a professor of civil, environmental and geodetic engineering, founding director of Ohio State’s Heart of Microbiome Science and a management group member within the EMERGE Biology Integration Institute.
“We’ve created a computationally reproducible technique to align these sequences to the place we will be extra assured that we're aligning positions that precisely mirror evolution.”
Additional evaluation utilizing 3D representations of sequence buildings and alignment revealed that the cluster of 5,500 new species didn’t match into the 5 present phyla of RNA viruses categorized within the Orthornavirae kingdom.
“We benchmarked our clusters in opposition to established, acknowledged phylogeny-based taxa, and that's how we discovered now we have extra clusters than people who existed,” mentioned co-first writer Ahmed Zayed, a analysis scientist in microbiology at Ohio State and a analysis lead within the EMERGE Institute.
In all, the findings led the researchers to suggest not solely the 5 new phyla, but in addition no less than 11 new orthornaviran courses of RNA viruses. The group is making ready a proposal to request formalization of the candidate phyla and courses by the ICTV.
Zayed mentioned the extent of latest knowledge on the RdRp gene’s divergence over time results in a greater understanding about how formative years could have advanced on the planet.
“RdRp is meant to be one of the crucial historical genes – it existed earlier than there was a necessity for DNA,” he mentioned. “So we’re not simply tracing the origins of viruses, but in addition tracing the origins of life.”
Reference: “Cryptic and considerable marine viruses on the evolutionary origins of Earth’s RNA virome” by Ahmed A. Zayed, James M. Wainaina, Guillermo Dominguez-Huerta, Eric Pelletier, Jiarong Guo, Mohamed Mohssen, Funing Tian, Akbar Adjie Pratama, Benjamin Bolduc, Olivier Zablocki, Dylan Cronin, Lindsey Solden, Erwan Delage, Adriana Alberti, Jean-Marc Aury, Quentin Carradec, Corinne da Silva, Karine Labadie, Julie Poulain, Hans-Joachim Ruscheweyh, Guillem Salazar, Elan Shatoff, Tara Oceans Coordinators, Ralf Bundschuh, Kurt Fredrick, Laura S. Kubatko, Samuel Chaffron, Alexander I. Culley, Shinichi Sunagawa, Jens H. Kuhn, Patrick Wincker, Matthew B. Sullivan, Silvia G. Acinas, Marcel Babin, Peer Bork, Emmanuel Boss, Chris Bowler, Man Cochrane, Colomban de Vargas, Gabriel Gorsky, Lionel Guidi, Nigel Grimsley, Pascal Hingamp, Daniele Iudicone, Olivier Jaillon, Stefanie Kandels, Lee Karp-Boss, Eric Karsenti, Fabrice Not, Hiroyuki Ogata, Nicole Poulton, Stéphane Pesant, Christian Sardet, Sabrinia Speich, Lars Stemmann, Matthew B. Sullivan, Shinichi Sungawa and Patrick Wincker, 7 April 2022, Science.
DOI: 10.1126/science.abm5847
This analysis was supported by the Nationwide Science Basis, the Gordon and Betty Moore Basis, the Ohio Supercomputer Heart, Ohio State’s Heart of Microbiome Science, the EMERGE Biology Integration Institute, the Ramon-Areces Basis and Laulima Authorities Options/NIAID. The work was additionally made attainable by the unprecedented sampling and science of the Tara Oceans Consortium, the nonprofit Tara Ocean Basis and its companions.
Extra co-authors on the paper have been co-lead authors James Wainaina and Guillermo Dominguez-Huerta, in addition to Jiarong Guo, Mohamed Mohssen, Funing Tian, Adjie Pratama, Ben Bolduc, Olivier Zablocki, Dylan Cronin and Lindsay Solden, all of Sullivan’s lab; Ralf Bundschuh, Kurt Fredrick, Laura Kubatko and Elan Shatoff of Ohio State’s School of Arts and Sciences; Hans-Joachim Ruscheweyh, Guillem Salazar and Shinichi Sunagawa of the Institute of Microbiology and Swiss Institute of Bioinformatics; Jens Kuhn of the Nationwide Institute of Allergy and Infectious Illnesses; Alexander Culley of the Université Laval; Erwan Delage and Samuel Chaffron of the Université de Nantes; and Eric Pelletier, Adriana Alberti, Jean-Marc Aury, Quentin Carradec, Corinne da Silva, Karine Labadie, Julie Poulain and Patrick Wincker of Genoscope.
Post a Comment