Quite a lot of current strains of SARS-CoV-2, in addition to different future variants that might come up, have the potential to flee the immune system’s cytotoxic T cell response in some portion of the inhabitants. That’s the conclusion of a brand new modeling research revealed on February 10th, 2022, in PLOS Computational Biology by Antonio Martín-Galiano of the Carlos III Well being Institute, Spain, and colleagues.
The T cell response in people is genetically encoded by HLA molecules—this implies totally different people have totally different HLAs, programmed to acknowledge invading pathogens primarily based on totally different elements, or “epitopes” of the pathogens. With hundreds of various HLA molecules within the human inhabitants and hundreds of doable epitopes in any given virus, the experimental analysis of the immune response of each human HLA allele to each viral variant shouldn't be possible. Nevertheless, computational strategies can facilitate this activity.
Within the new research, researchers first decided the complete set of epitopes from an unique reference pressure of SARS-CoV-2 from Wuhan, China. The staff found 1,222 epitopes of SARS-CoV-2 that have been related to main HLA subtypes, overlaying about 90% of the human inhabitants; at the very least 9 out of each 10 individuals can launch a T cell response to COVID-19 primarily based on these 1,222 epitopes.
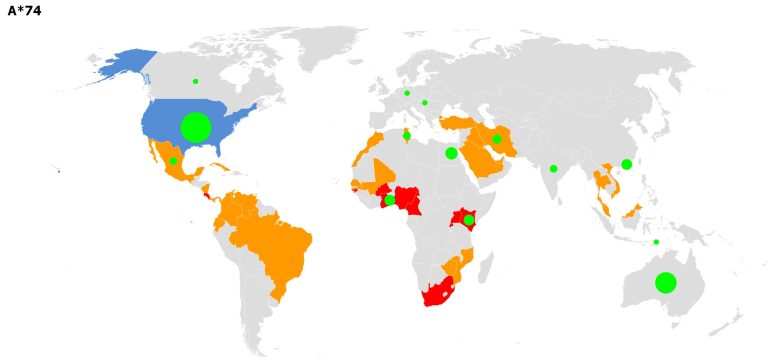
World map indicating the presence of inhabitants samples carrying alleles of the A*74 household and isolates with escape mutations for these household (inexperienced circles). Credit score: Foix A et al., 2022, PLOS Computational Biology, CC-BY 4.0
Then, the researchers computationally analyzed whether or not any of 118,000 totally different SARS-CoV-2 isolates from around the globe, described in a Nationwide Heart for Biotechnology Info (NCBI) dataset, had mutations in these epitopes. 47% of the epitopes, they confirmed, have been mutated in at the very least one current isolate. In some instances, current isolates had mutations in a number of epitope areas, however cumulative mutations by no means affected greater than 15% of epitopes for any given HLA allele kind. When the staff analyzed vulnerable alleles and the geographic origin of their respective escape isolates, the staff discovered that they co-existed in some geographical areas—together with sub-Saharan Africa and East and Southeast Asia—suggesting potential genetic strain on the cytotoxic T cell response in these areas.
“The buildup of those adjustments in impartial isolates remains to be too low to threaten the worldwide human inhabitants,” the authors say. “Our protocol has recognized mutations which may be related for particular populations and warrant deeper surveillance.”
Nevertheless, Martín-Galiano notes that “unnoticed SARS-CoV-2 mutations” may sooner or later “threaten the cytotoxic T response in human subpopulations.”
Reference: “Predicted impression of the viral mutational panorama on the cytotoxic response in opposition to SARS-CoV-2” by Anna Foix, Daniel López, Francisco Díez-Fuertes,
Michael J. McConnell and Antonio J. Martín-Galiano, 10 February 2022, PLoS Computational Biology.
DOI: 10.1371/journal.pcbi.1009726
Funding: This analysis was supported by Acción Estratégica en Salud from the ISCIII, grants MPY 380/18 (to MJM), 388/18 (to DL) and 509/19 (to AJM-G). AJM-G is the recipient of a Miguel Servet contract by the ISCIII. The funders had no function in research design, knowledge assortment and evaluation, choice to publish, or preparation of the manuscript.
Post a Comment