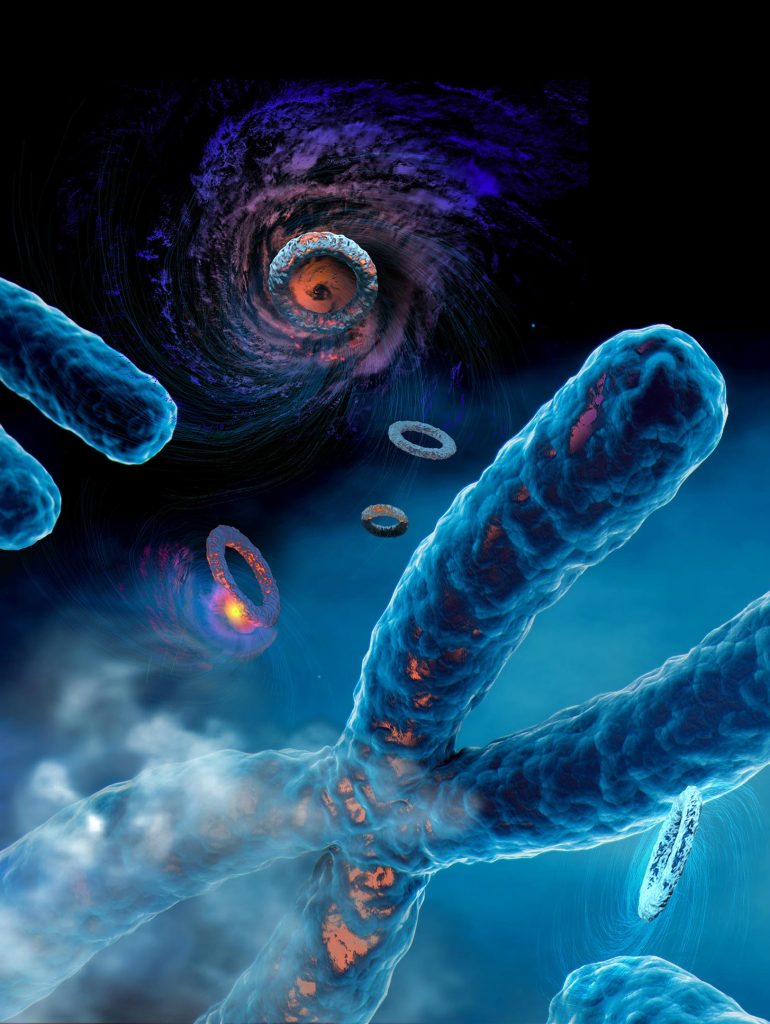
This creative rendering illustrates the variety of mutational processes that generate clustered mutations in human most cancers. Depicted listed here are kyklonas, that are molecular cyclones that trigger mutations on round extrachromosomal DNA (ecDNA), and omikli, which is a molecular fog that causes mutations on linear chromosomal DNA. Credit score: Catherine Eng
Researchers led by bioengineers on the College of California San Diego have recognized and characterised a beforehand unrecognized key participant in most cancers evolution: clusters of mutations occurring at sure areas of the genome. The researchers discovered that these mutation clusters contribute to the development of about 10% of human cancers and can be utilized to foretell affected person survival.
The findings have been reported in a paper revealed on February 9, 2022, within the journal Nature.
The work sheds gentle on a category of mutations referred to as clustered somatic mutations—clustered which means they group collectively at particular areas in a cell’s genome, and somatic which means they don't seem to be inherited, however attributable to inner and exterior elements corresponding to growing older or publicity to UV radiation, for instance.
Clustered somatic mutations have to this point been an understudied space in most cancers growth. However researchers within the lab of Ludmil Alexandrov, a professor of bioengineering and mobile and molecular drugs at UC San Diego, noticed one thing extremely uncommon about these mutations that warranted additional research.
“We usually see somatic mutations occurring randomly throughout the genome. However after we appeared nearer at a few of these mutations, we noticed that they have been occurring in these hotspots. It’s like throwing balls on the ground after which abruptly seeing them cluster in a single area,” mentioned Alexandrov. “So we couldn’t assist however surprise: What is occurring right here? Why are there hotspots? Are they clinically related? Do they inform us one thing about how most cancers has developed?”
“Clustered mutations have largely been ignored as a result of they solely make up a really small proportion of all mutations,” mentioned Erik Bergstrom, a bioengineering PhD scholar in Alexandrov’s lab and the primary writer of the research. “However by diving deeper, we discovered that they play an essential position within the etiology of human most cancers.”
The crew’s discoveries have been enabled by creating essentially the most complete and detailed map of recognized clustered somatic mutations. They began by mapping all of the mutations (clustered and non-clustered) throughout the genomes of greater than 2500 most cancers sufferers—an effort that in whole encompassed 30 totally different most cancers sorts. The researchers created their map utilizing next-generation synthetic intelligence approaches developed within the Alexandrov lab. The crew used these algorithms to detect clustered mutations inside particular person sufferers and elucidate the underlying mutational processes that give rise to such occasions. This led to their discovering that clustered somatic mutations contribute to most cancers evolution in roughly 10% of human cancers.
Taking it a step additional, the researchers additionally discovered that a number of the cancer-driving clusters—particularly these present in recognized most cancers driver genes—can be utilized to foretell the general survival of a affected person. For instance, the presence of clustered mutations within the BRAF gene—essentially the most broadly noticed driver gene in melanoma—ends in higher total affected person survival in comparison with people with non-clustered mutations. In the meantime, the presence of clustered mutations within the EGFR gene—essentially the most broadly noticed driver gene in lung most cancers—ends in decreased affected person survival.
“What’s attention-grabbing is that we see differential survival by way of simply having clustered mutations detected inside these genes, and that is detectable with current platforms which are generally used within the clinic. So this acts as a quite simple and exact biomarker for affected person survival,” mentioned Bergstrom.
“This elegant work emphasizes the significance of growing AI approaches to elucidate tumor biology, and for biomarker discovery and speedy growth utilizing normal platforms with direct line of sight translation to the clinic,” mentioned Scott Lippman, director of Moores Most cancers Heart and affiliate vice chancellor for most cancers analysis and care at UC San Diego. “This highlights UC San Diego’s power in combining engineering approaches in synthetic intelligence for fixing present issues in most cancers drugs.”
A brand new mode of most cancers evolution
On this research, the researchers additionally recognized numerous elements that trigger clustered somatic mutations. These elements embody UV radiation, alcohol consumption, tobacco smoking, and most notably, the exercise of a set of antiviral enzymes referred to as APOBEC3.
APOBEC3 enzymes are usually discovered inside cells as a part of their inner immune response. Their principal job is to cut up any viruses that enter the cell. However in most cancers cells, the researchers assume that the APOBEC3 enzymes could also be doing extra hurt than good.
The researchers discovered that most cancers cells—which are sometimes rife with round rings of extrachromosomal DNA (ecDNA) that harbor recognized most cancers driver genes—have clusters of mutations occurring throughout particular person ecDNA molecules. The researchers attribute these mutations to the exercise of APOBEC3 enzymes. They hypothesize that APOBEC3 enzymes are mistaking the round rings of ecDNA as overseas viruses and try to limit and chop them up. In doing so, the APOBEC3 enzymes trigger clusters of mutations to kind inside particular person ecDNA molecules. This in flip performs a key position in accelerating most cancers evolution and certain results in drug resistance. The researchers named these rings of clustered mutations kyklonas, which is the Greek phrase for cyclones.
“It is a utterly novel mode of oncogenesis,” mentioned Alexandrov. Together with the crew’s different findings, he defined, “this lays the inspiration for brand new therapeutic approaches, the place clinicians can take into account proscribing the exercise of APOBEC3 enzymes and/or focusing on extrachromosomal DNA for most cancers therapy.”
Reference: “Mapping clustered mutations in most cancers reveals APOBEC3 mutagenesis of ecDNA” by Erik N. Bergstrom, Jens Luebeck, Mia Petljak, Azhar Khandekar, Mark Barnes, Tongwu Zhang, Christopher D. Steele, Nischalan Pillay, Maria Teresa Landi, Vineet Bafna, Paul S. Mischel, Reuben S. Harris and Ludmil B. Alexandrov, 9 February 2022, Nature.
DOI: 10.1038/s41586-022-04398-6
This work was supported by a Most cancers Grand Problem award from Most cancers Analysis UK in addition to funding from the U.S. Nationwide Institutes of Well being, Alfred P. Sloan Basis, and Packard Basis.
Post a Comment